Human bone marrow of healthy human donors#
In this notebook, we pre-process cyTOF data of bone marrow samples from 8 healthy donors. Data were provided by Oetjen et al (JCl Insight, 2018). We employ the following steps:
Load and convert fcs file into anndata format
Arcsinh-normalisation with pre-determined cofactor
Compute knn-graph
Compute Leiden clustering
In the next notebook, we focus on the annotation of the clusters and compare with the original annotation provided by the authors.
import scanpy as sc
import anndata as ann
import numpy as np
import scipy as sp
import pandas as pd
import matplotlib.pyplot as pl
from matplotlib import rcParams
from matplotlib import colors
import seaborn as sb
import datetime
import pytometry as pm
sc.logging.print_versions()
sc.settings.verbosity = 3
WARNING: If you miss a compact list, please try `print_header`!
The `sinfo` package has changed name and is now called `session_info` to become more discoverable and self-explanatory. The `sinfo` PyPI package will be kept around to avoid breaking old installs and you can downgrade to 0.3.2 if you want to use it without seeing this message. For the latest features and bug fixes, please install `session_info` instead. The usage and defaults also changed slightly, so please review the latest README at https://gitlab.com/joelostblom/session_info.
-----
anndata 0.7.6
scanpy 1.8.1
sinfo 0.3.4
-----
PIL 8.4.0
anyio NA
asciitree NA
attr 21.2.0
babel 2.9.1
backcall 0.2.0
beta_ufunc NA
binom_ufunc NA
brotli 1.0.9
certifi 2021.10.08
cffi 1.15.0
charset_normalizer 2.0.7
cloudpickle 2.0.0
cycler 0.10.0
cython_runtime NA
dask 2021.10.0
dateutil 2.8.2
debugpy 1.5.1
decorator 5.1.0
defusedxml 0.7.1
entrypoints 0.3
fasteners 0.17.3
flowio 1.0.1
fsspec 2022.3.0
google NA
h5py 3.1.0
idna 3.3
igraph 0.9.7
ipykernel 6.4.2
ipython_genutils 0.2.0
ipywidgets 7.6.5
jedi 0.18.0
jinja2 3.0.2
joblib 1.1.0
json5 NA
jsonschema 4.1.2
jupyter_server 1.11.1
jupyterlab_server 2.8.2
kiwisolver 1.3.2
leidenalg 0.8.8
llvmlite 0.37.0
louvain 0.7.0
markupsafe 2.0.1
matplotlib 3.4.3
matplotlib_inline NA
mpl_toolkits NA
msgpack 1.0.3
natsort 7.1.1
nbclassic NA
nbformat 5.1.3
nbinom_ufunc NA
numba 0.54.1
numcodecs 0.9.1
numexpr 2.7.3
numpy 1.19.5
packaging 21.0
pandas 1.3.4
parso 0.8.2
pexpect 4.8.0
pickleshare 0.7.5
pkg_resources NA
prometheus_client NA
prompt_toolkit 3.0.21
ptyprocess 0.7.0
pvectorc NA
pydev_ipython NA
pydevconsole NA
pydevd 2.6.0
pydevd_concurrency_analyser NA
pydevd_file_utils NA
pydevd_plugins NA
pydevd_tracing NA
pygments 2.10.0
pyparsing 3.0.1
pyrsistent NA
pytometry 0.0.1
pytz 2021.3
readfcs 0.1.5
requests 2.26.0
scipy 1.7.1
seaborn 0.11.2
send2trash NA
setuptools_scm NA
six 1.15.0
sklearn 1.0.1
sniffio 1.2.0
socks 1.7.1
sparse 0.13.0
statsmodels 0.13.0
storemagic NA
tables 3.6.1
terminado 0.12.1
texttable 1.6.4
threadpoolctl 3.0.0
tlz 0.11.1
toolz 0.11.1
tornado 6.1
traitlets 5.1.1
typing_extensions NA
urllib3 1.26.7
wcwidth 0.2.5
websocket 1.2.1
yaml 6.0
zarr 2.11.3
zmq 22.3.0
-----
IPython 7.28.0
jupyter_client 7.0.6
jupyter_core 4.8.1
jupyterlab 3.2.1
notebook 6.4.5
-----
Python 3.8.6 (default, Oct 26 2021, 09:26:31) [GCC 8.3.0]
Linux-4.18.0-305.12.1.el8_4.x86_64-x86_64-with-glibc2.28
288 logical CPU cores
-----
Session information updated at 2022-08-11 12:11
Add date.
now = datetime.datetime.now()
today = now.strftime("%Y%m%d")
# Define a nice colour map for gene expression
colors2 = pl.cm.Reds(np.linspace(0, 1, 80))
colors3 = pl.cm.Greys_r(np.linspace(0.7, 0.8, 35))
colorsComb = np.vstack([colors3, colors2])
mymap = colors.LinearSegmentedColormap.from_list("my_colormap", colorsComb)
import os
data_path = "./../data/Oetjen_2018/"
Read data#
files_all = os.listdir(data_path + "cytof_data/")
files = [fileID for fileID in files_all if fileID.endswith(".fcs")]
files
['20171103_T_01_1.fcs',
'20171103_U_01_1.fcs',
'20171103_B_01_1.fcs',
'20171103_A_01_1.fcs',
'20171103_H_01_1.fcs',
'20171103_C_01_1.fcs',
'20171103_O_01_1.fcs',
'20171103_J_01_1.fcs']
adatas = []
for fileID in files:
meta_info = fileID.split(".fcs")[0].split("_")
adata = pm.io.read_fcs(data_path + "cytof_data/" + fileID)
adata.obs["sample"] = meta_info[1]
# move Time etc to .obs
pm.pp.split_signal(adata, var_key="channel", option="element", data_type="cytof")
adatas.append(adata)
adatas[0].var_names
Index(['89Y-CD45', '103Rh', '120Sn-Environ', '127I', '131Xe-Environ',
'133Cs-Environ', '138Ba-Environ', '140Ce-EQ4-beads', '141Pr-CD49D',
'142Nd-CD11a', '143Nd-CD5', '144Nd-CD195', '145Nd-CD4', '146Nd-CD8a',
'147Sm-CD7', '148Nd-CD16', '149Sm-CD25', '150Nd-CD134---OX40',
'151Eu-CD2', '152Sm-CD95---FAS', '153Eu-CD366---TIM3', '154Sm',
'155Gd-CD279---PD1', '156Gd-CD183', '158Gd-CD194', '159Tb-CD197',
'160Gd-CD28', '161Dy-CD152---CTLA4', '162Dy-CD69', '163Dy',
'164Dy-CD161', '165Ho-CD45RO', '166Er-CD44', '167Er-CD27',
'168Er-CD278---ICOS', '169Tm-CD45RA', '170Er-CD3', '171Yb-CD9',
'172Yb-CD57', '173Yb-CD137---41BB', '174Yb-HLA-DR',
'175Lu-CD223---LAG3', '176Yb-CD127', '190BCKG', '191Ir-DNA1',
'193Ir-DNA2', '195Pt-VIABILITY', '208Pb-Environ', '209Bi'],
dtype='object')
Concatenate all data.
adata_all = ann.AnnData.concatenate(*adatas, join="outer", uns_merge="unique")
Remove unused channels. According to Supplementary Table S5 of the Oetjen et al. publication, less channels were used than reported in the file. In addition, we add the corresponding antibody marker for each element.
marker_list = adata_all.var["marker"].values
rename_dict = {}
for marker in marker_list:
elem_info = marker.split("-")
rename_dict[marker] = elem_info[-1] if len(elem_info) > 1 else "unused"
if elem_info[-1] == "DR": # fix HLR-DR
rename_dict[marker] = "-".join(elem_info[-2:])
adata_all.var["AB"] = pd.Categorical(adata_all.var["marker"]).map(rename_dict)
Remove the unused channels, i.e. the ones with no marker or the ones termed ‘Environ’ or ‘EQ4beads’.
marker_keep = [
marker
for marker in adata_all.var["AB"]
if marker not in ["unused", "Environ", "beads"]
]
adata_all = adata_all[:, np.in1d(adata_all.var["AB"], marker_keep)]
adata_all
View of AnnData object with n_obs × n_vars = 4938816 × 37
obs: 'sample', 'Time', 'Event-length', 'Center', 'Offset', 'Width', 'Residual', 'batch'
var: 'channel', 'marker', 'signal_type', 'AB'
uns: 'meta'
Save to file.
adata_all.write(data_path + "anndata/" + "cytof_data_concatenated.h5ad")
/opt/python/lib/python3.8/site-packages/anndata/_core/anndata.py:1220: FutureWarning: The `inplace` parameter in pandas.Categorical.reorder_categories is deprecated and will be removed in a future version. Reordering categories will always return a new Categorical object.
c.reorder_categories(natsorted(c.categories), inplace=True)
/opt/python/lib/python3.8/site-packages/anndata/_core/anndata.py:1228: ImplicitModificationWarning: Initializing view as actual.
warnings.warn(
Trying to set attribute `.obs` of view, copying.
... storing 'sample' as categorical
Preprocess data#
adata_all = sc.read(data_path + "anndata/" + "cytof_data_concatenated.h5ad")
adata_all
AnnData object with n_obs × n_vars = 4938816 × 37
obs: 'sample', 'Time', 'Event-length', 'Center', 'Offset', 'Width', 'Residual', 'batch'
var: 'channel', 'marker', 'signal_type', 'AB'
uns: 'meta'
Check sample size.
adata_all.obs["sample"].value_counts()
B 956776
O 934338
H 848865
J 599766
T 517921
A 500671
C 381121
U 199358
Name: sample, dtype: int64
Normalise data#
We use a normalization cofactor of 5.
cofactor = 5
Save original data as layer.
adata_all.layers["original"] = adata_all.X
Normalize.
pm.tl.normalize_arcsinh(adata=adata_all, cofactor=cofactor)
AnnData object with n_obs × n_vars = 4938816 × 37
obs: 'sample', 'Time', 'Event-length', 'Center', 'Offset', 'Width', 'Residual', 'batch'
var: 'channel', 'marker', 'signal_type', 'AB'
uns: 'meta'
layers: 'original'
Filter for viability and DNA staining#
The data comes with three markers for DNA content and viability staining. Let us investigate the data quality based on these markers and filter out potentially dead cells.
rcParams["figure.figsize"] = (7, 7)
sc.pl.scatter(adata_all, x="191Ir-DNA1", y="193Ir-DNA2", color="sample", size=2)
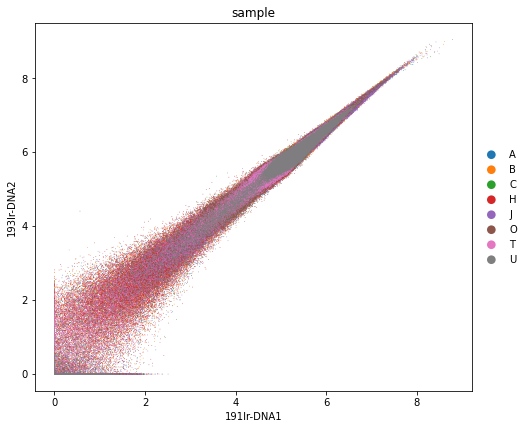
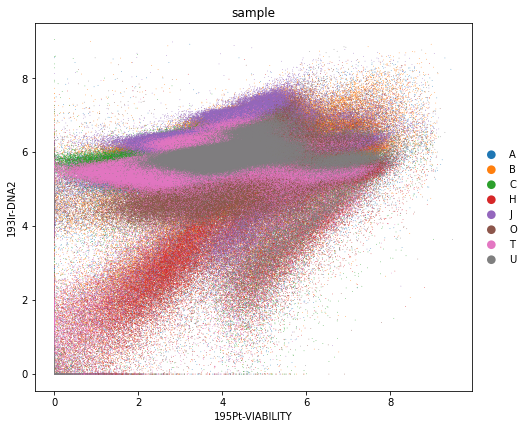
rcParams["figure.figsize"] = (7, 7)
sc.pl.scatter(adata_all, x="195Pt-VIABILITY", y="193Ir-DNA2", color="sample", size=2)
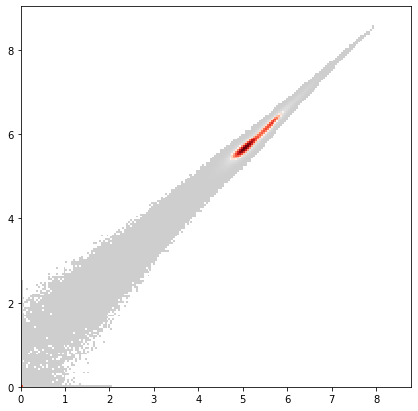
x = adata_all.var["AB"] == "DNA1"
y = adata_all.var["AB"] == "DNA2"
ax = pl.hist2d(
adata_all.X[:, x].flatten(),
adata_all.X[:, y].flatten(),
bins=200,
cmin=5,
cmap=mymap,
)
pl.show()
x = adata_all.var["AB"] == "VIABILITY"
y = adata_all.var["AB"] == "DNA1"
ax = pl.hist2d(
adata_all.X[:, x].flatten(),
adata_all.X[:, y].flatten(),
bins=200,
cmin=5,
cmap=mymap,
)
pl.show()

Overall, we consider all events with less than 2 in either ‘DNA1’, ‘DNA2’ or less than 1 in ‘VIABILITY’ as poor quality events and filter them out.
dna1_low = adata_all.X[:, adata_all.var["AB"] == "DNA1"].flatten() < 2
dna2_low = adata_all.X[:, adata_all.var["AB"] == "DNA2"].flatten() < 2
viab_low = adata_all.X[:, adata_all.var["AB"] == "VIABILITY"].flatten() < 1
viability_filter = (dna1_low + dna2_low + viab_low) == 0
np.sum(viability_filter)
4829382
adata_all = adata_all[viability_filter]
Remove ‘DNA1’, ‘DNA2’ and ‘VIABILITY’ from data matrix.
viab_marker = ["DNA1", "DNA2", "VIABILITY"]
for marker in viab_marker:
adata_all.obs[marker] = adata_all.X[:, adata_all.var["AB"] == marker]
adata_all = adata_all[:, np.invert(np.in1d(adata_all.var["AB"], viab_marker))].copy()
Trying to set attribute `.obs` of view, copying.
adata_all
AnnData object with n_obs × n_vars = 4829382 × 34
obs: 'sample', 'Time', 'Event-length', 'Center', 'Offset', 'Width', 'Residual', 'batch', 'DNA1', 'DNA2', 'VIABILITY'
var: 'channel', 'marker', 'signal_type', 'AB'
uns: 'meta', 'sample_colors'
layers: 'compensated'
Save normalised and filtered data to file.
adata_all.write(data_path + "anndata/" + "cytof_data_norm.h5ad")
Visualise data as UMAP#
adata_all = sc.read(data_path + "anndata/" + "cytof_data_norm.h5ad")
In the next step, we compute a knn-graph, an embedding and a clustering of the data.
sc.pp.pca(adata_all)
computing PCA
with n_comps=33
finished (0:00:13)
sc.pl.pca_overview(adata_all, color="sample")


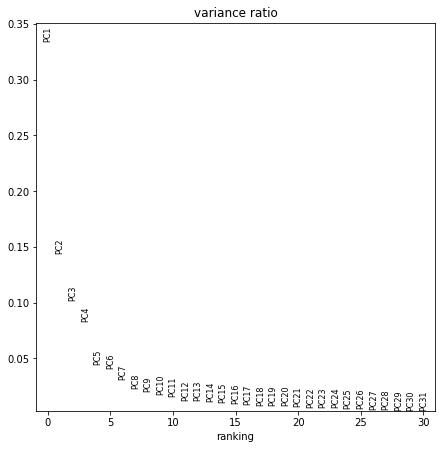
sc.pp.pca(adata_all, n_comps=10)
sc.pp.neighbors(adata_all, n_neighbors=15, use_rep="X_pca")
computing PCA
with n_comps=10
finished (0:00:10)
computing neighbors
finished: added to `.uns['neighbors']`
`.obsp['distances']`, distances for each pair of neighbors
`.obsp['connectivities']`, weighted adjacency matrix (0:18:48)
adata_all
AnnData object with n_obs × n_vars = 4829382 × 34
obs: 'sample', 'Time', 'Event-length', 'Center', 'Offset', 'Width', 'Residual', 'batch', 'DNA1', 'DNA2', 'VIABILITY'
var: 'channel', 'marker', 'signal_type', 'AB'
uns: 'meta', 'sample_colors', 'pca', 'neighbors', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'compensated'
obsp: 'distances', 'connectivities'
adata_all.write(data_path + "anndata/" + "cytof_data_norm.h5ad")
adata_all = sc.read(data_path + "anndata/" + "cytof_data_norm.h5ad")
adata_all
AnnData object with n_obs × n_vars = 4829382 × 34
obs: 'sample', 'Time', 'Event-length', 'Center', 'Offset', 'Width', 'Residual', 'batch', 'DNA1', 'DNA2', 'VIABILITY'
var: 'channel', 'marker', 'signal_type', 'AB'
uns: 'meta', 'sample_colors', 'pca', 'neighbors', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'compensated'
obsp: 'distances', 'connectivities'
sc.tl.umap(adata_all)
computing UMAP
sc.pl.umap(adata_all, color="sample")

Save to file#
adata_all.write(data_path + "anndata/" + "cytof_data_norm.h5ad")
End of the pre-processing notebook.